A New Weapon for Tuberculosis Diagnosis and Drug Resistance Detection: A New Generation Targeted Sequencing (tNGS) Combined with Machine Learning for Tuberculosis Hypersensitivity Diagnosis
Literature report: CCa: a diagnostic model based on tNGS and machine learning, which is suitable for people with less bacterial tuberculosis and tuberculous meningitis.
Thesis title: Tuberculous-targeted next-generation sequencing and machine learning: an ultra-sensitive diagnostic strategy for paucific pulmonary tubulars and tubular meningitis.
Periodical: 《Clinica Chimica Acta》
IF:6.5
Date of publication: January 2024
Combined with the University of Chinese Academy of Sciences and Beijing Chest Hospital of Capital Medical University, Macro & Micro-Test established a tuberculosis diagnosis model based on the new generation of targeted sequencing (tNGS) technology and machine learning method, which provided ultra-high detection sensitivity for tuberculosis with few bacteria and tuberculous meningitis, provided a new hypersensitivity diagnosis method for the clinical diagnosis of two types of tuberculosis, and helped the accurate diagnosis, drug resistance detection and treatment of tuberculosis. At the same time, it is found that the patient's plasma cfDNA can be used as a suitable sample type for clinical sampling in the diagnosis of TBM.
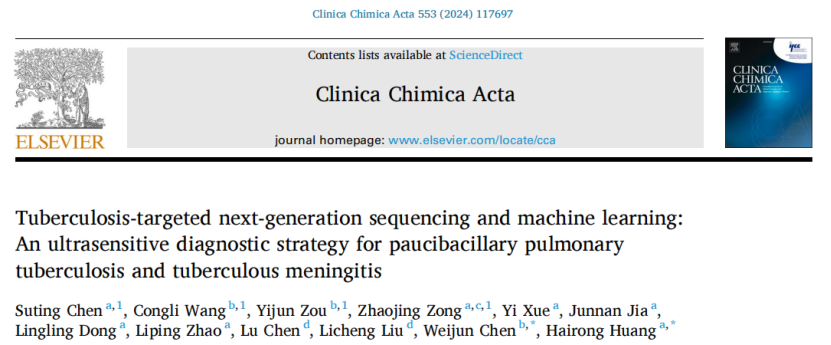
In this study, 227 plasma samples and cerebrospinal fluid samples were used to establish two clinical cohorts, in which the laboratory diagnostic cohort samples were used to establish the machine learning model of tuberculosis diagnosis, and the clinical diagnostic cohort samples were used to verify the established diagnostic model. All samples were first targeted by a specially designed targeted capture probe pool for Mycobacterium tuberculosis. Then, based on TB-tNGS sequencing data, the decision tree model is used to perform 5-fold cross-validation on the training and validation sets of the laboratory diagnostic queue, and the diagnostic thresholds of plasma samples and cerebrospinal fluid samples are obtained. The obtained threshold is brought into two test sets of clinical diagnosis queue for detection, and the diagnostic performance of the learner is evaluated by ROC curve. Finally, the diagnosis model of tuberculosis was obtained.
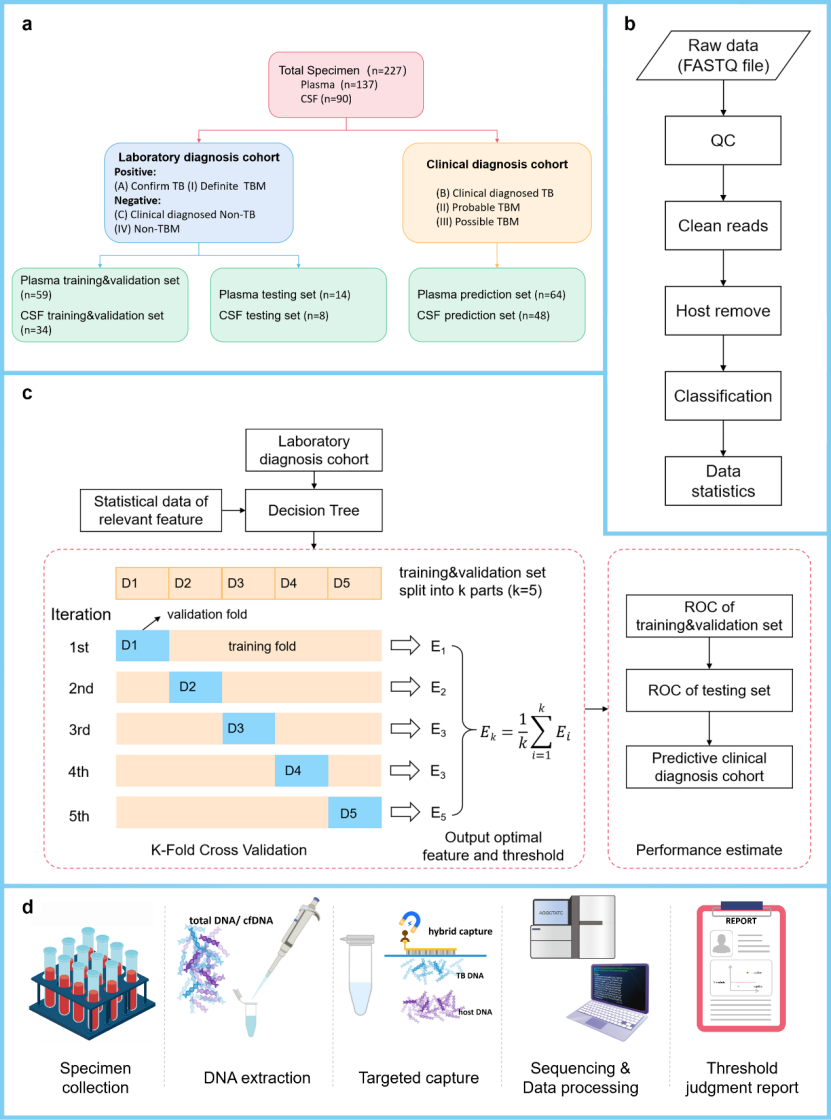
Fig. 1 schematic diagram of research design
Results: According to the specific thresholds of CSF DNA sample (AUC = 0.974) and plasma cfDNA sample (AUC = 0.908) determined in this study, among 227 samples, the sensitivity of CSF sample was 97.01%, the specificity was 95.65%, and the sensitivity and specificity of plasma sample were 82.61% and 86.36%. In the analysis of 44 paired samples of plasma cfDNA and cerebrospinal fluid DNA from TBM patients, the diagnostic strategy of this study has a high consistency of 90.91% (40/44) in plasma cfDNA and cerebrospinal fluid DNA, and the sensitivity is 95.45% (42/44). In children with pulmonary tuberculosis, the diagnostic strategy of this study is more sensitive to plasma samples than the Xpert detection results of gastric juice samples from the same patients (28.57% VS 15.38%).
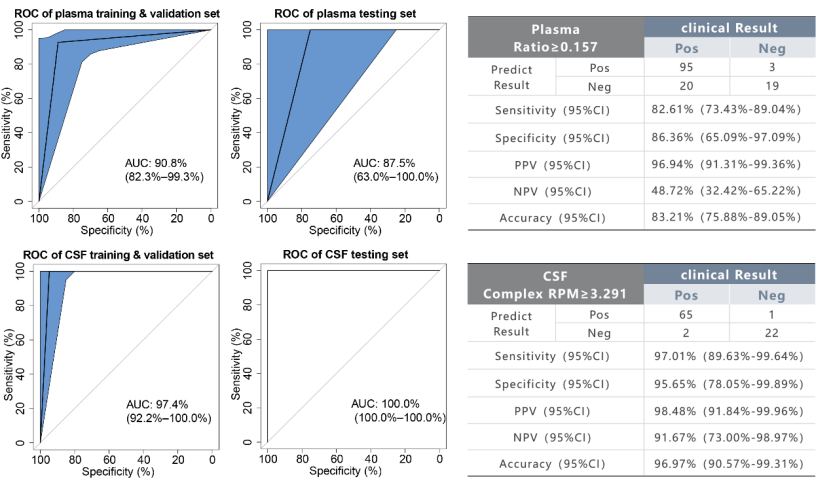
Fig. 2 Analysis performance of tuberculosis diagnosis model for population samples
Fig. 3 Diagnostic results of paired samples
Conclusion: A hypersensitive diagnostic method for tuberculosis was established in this study, which can provide a diagnostic tool with the highest detection sensitivity for clinical patients with oligobacillary tuberculosis (negative culture). Detection of hypersensitive tuberculosis based on plasma cfDNA may be a suitable sample type for the diagnosis of active tuberculosis and tuberculous meningitis (plasma samples are easier to collect than cerebrospinal fluid for patients suspected of brain tuberculosis).
Original link: https://www.sciencedirect.com/science/article/pii/s0009898123004990? via%3Dihub
Brief introduction of Macro & Micro-Test tuberculosis series detection products
In view of the complicated sample situation of tuberculosis patients and various needs, Macro & Micro-Test provides a complete set of NGS solutions for liquefaction extraction from sputum samples, Qualcomm library construction and sequencing, and data analysis. The products cover rapid diagnosis of tuberculosis patients, drug resistance detection of tuberculosis, typing of mycobacterium tuberculosis and NTM, hypersensitivity diagnosis of bacteria-negative tuberculosis and tuberculous people, etc.
Serial detection kits for tuberculosis and mycobacteria:
| Item No | product name | product testing content | sample type | applicable model |
| HWTS-3012 | Sample release agent | Used in the liquefaction treatment of sputum samples, has obtained a first-class record number, Sutong Machinery Equipment 20230047. | sputum | |
| HWTS-NGS-P00021 | Qualcomm quantity detection kit for hypersensitive tuberculosis (probe capture method) | Non-invasive (liquid biopsy) hypersensitivity detection for bacteria-negative pulmonary tuberculosis and brain nodules; The samples of people suspected to be infected with tuberculosis or non-tuberculosis mycobacteria were analyzed by high-depth sequencing metagenomics, and the detection information of whether tuberculosis or non-tuberculosis mycobacteria were infected and the main first-line drug resistance information of mycobacterium tuberculosis were provided. | Peripheral blood, alveolar lavage fluid, hydrothorax and ascites, focus puncture sample, cerebrospinal fluid. | Second generation |
| HWTS-NGS-T001 | Mycobacterium typing and drug resistance detection kit (multiplex amplification sequencing method) | Mycobacterium typing test, including MTBC and 187 NTM;Drug resistance detection of Mycobacterium tuberculosis covers 13 drugs and 16 core mutation sites of drug resistance genes. | Sputum, alveolar lavage fluid, hydrothorax and ascites, focus puncture sample, cerebrospinal fluid. | Second/third generation dual platform |
Highlights: HWTS-NGS-T001 Mycobacterium typing and drug resistance detection kit (multiplex amplification method)
Product introduction
The product is based on the main first-and second-line drugs described in the WHO TB treatment guidelines, macrolides and aminoglycosides commonly used in the NTM treatment guidelines, and the drug resistance sites cover all one group of drug resistance-related sites in the WHO Mycobacterium tuberculosis complex mutation catalogue, as well as other reported drug resistance genes and mutation sites according to the investigation and statistics of high-scoring literatures at home and abroad.
Typing identification is based on the NTM strains summarized in the NTM guidelines published by Chinese Journal of Tuberculosis and Respiratory Diseases and the consensus of experts. The designed typing primers can amplify, sequence and annotate more than 190 NTM species.
Through targeted multiplex PCR amplification technology, the genotyping genes and drug-resistant genes of Mycobacterium were amplified by multiplex PCR, and the amplicon combination of target genes to be detected was obtained. The amplified products can be constructed into second-generation or third-generation high-throughput sequencing libraries, and all second-generation and third-generation sequencing platforms can be subjected to high-depth sequencing to obtain the sequence information of target genes. By comparing with the known mutations contained in the built-in reference database (including the WHO Mycobacterium tuberculosis complex mutation catalogue and its relationship with drug resistance), the mutations related to drug resistance or susceptibility of anti-tuberculosis drugs were determined. Combined with the self-opened sputum sample treatment solution of Macro & Micro-Test, the problem of low nucleic acid amplification efficiency of clinical sputum samples (ten times higher than that of traditional methods) was solved, so that drug resistance sequencing detection can be directly applied to clinical sputum samples.
Product detection range
34 drug resistance-related genes of 18 anti-tuberculosis drugs and 6 NTM drugs were detected, covering 297 drug resistance sites; Ten kinds of Mycobacterium tuberculosis and more than 190 kinds of NTM were detected.
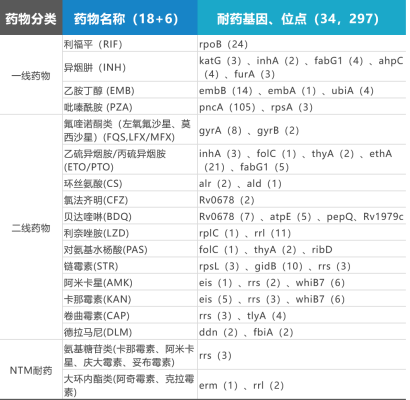
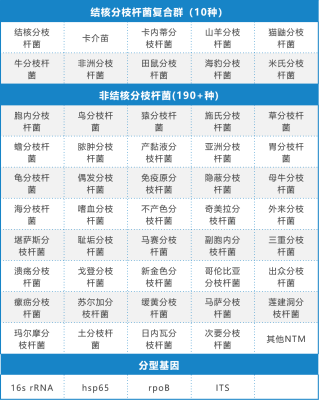
Table 1: Information of 18+6 Drugs +190+NTM
Product advantage
Strong clinical adaptability: the sputum samples can be directly detected with self-liquefaction agent without culture.
The experimental operation is simple: the first round of amplification operation is simple, and the library construction is completed in 3 hours, which improves the work efficiency.
Comprehensive typing and drug resistance: covering the typing and drug resistance sites of MTB and NTM, which are the key points of clinical concern, accurate typing and drug resistance detection, supporting independent analysis software, and generating analysis reports with one click.
Compatibility: product compatibility, adapting to mainstream ILM and MGI/ONT platforms.
Product specification
| Product code | product name | Detection platform | specifications |
| HWTS-NGS-T001 | Mycobacterium typing and drug resistance detection kit (multiplex amplification method) | ONT、Illumina、MGI、Salus pro | 16/96rxn |
Post time: Jan-23-2024
